 |
JASSA v4 Joined Advanced Sumoylation Site and Sim Analyser |
|
| by BEAUCLAIR Guillaume |
 |
 |
Analyser |
 |
Documentation |
 |
Contact |
 |
 |
|
Documentation in update
Comming soon... In update...
About SUMOylation
SUMOylation is a post-translational modification conserved from yeast to human that modulates several fundamental cellular processes, and have been showed to be involved in human disorders. It consists of the covalent attachment of a small ubiquitin-related modifier protein (SUMO) to a target protein by a mechanism similar to that of ubiquitination. (Melchior, 2000; Geiss-Friedlander and Melchior, 2007). There are at least three SUMO isoforms (SUMO1,2,3) in mammalian cells but only one in Saccharomyces cerevisiae (Smt3).
This post-translational modification involves a cascade of SUMO-specific enzymes (for a review, see (Johnson, 2004; Kerscher et al., 2006; Martin et al., 2007)). First, SUMO is proteolytically cleaved to expose the internal diglycine motif required for conjugation. Then SUMO is activated in an ATP-dependent manner by the heterodimeric SUMO activating enzyme SAE1/SAE2. SUMO is transferred to the conjugating enzyme Ubc9 and is conjugated to the target substrate protein. This process can be enhanced by involvement of growing number of E3 ligases. SUMO peptides is reversibly covalent conjugated onto an acceptor lysine residue (K) of the substrate which often lies within the consensus sequence ΨKxE (where Ψ is an hydrophobic residue and x any amino acid; Melchior, 2000; Rodriguez et al., 2001). It should be noted that inverted SUMOylation motif ([E/D]xKΨ) have been reported few years ago (Matic et al., 2010) (Table 1). Extended SUMO consensus motifs, like consensus inverted, PDSM (phosphorylation-dependent SUMOylation motif) (Gregoire et al., 2006; Hietakangas et al., 2006; Shalizi et al., 2006), NDSM (negatively charged amino acid-dependent SUMOylation site) (Yang et al., 2006), SUMO-acetyl switch (Stankovic-Valentin et al., 2007) and HCSM (hydrophobic cluster SUMOylation motif) (Matic et al., 2010; Hietakangas et al., 2006) also have been reported (for a review, see (Martin et al., 2007)). Moreover, elements flanking the core motif (such as acidic, phosphorylatable or proline residues) may impact on SUMO conjugation (Table 1) (Gareau and Lima, 2011). Notably, about 25% of experimentally validated SUMOylated sites do not match with any of these motifs. Additionally, not all sites that adhere to the consensus are modified, likely because SUMO is conjugated only to residues appropriately presented to the SUMOylation machinery. SUMO can also interact non-covalently with proteins harboring SUMO-interacting motifs (SIM), also known as SUMO-binding domains (SBDs) or motifs (SBMs), typically consisting of a hydrophobic core (Kerscher, 2007) (Table 1). Negatively charged patches of residues flanking the SIM may contribute to the orientation and/or the isoforme-specificity binding (Hannich et al., 2005; Hecker et al., 2006) and an implication of the phosphorylation near the SIM, named phosphoSIM, have been reported (Stehmeier and Muller, 2009). 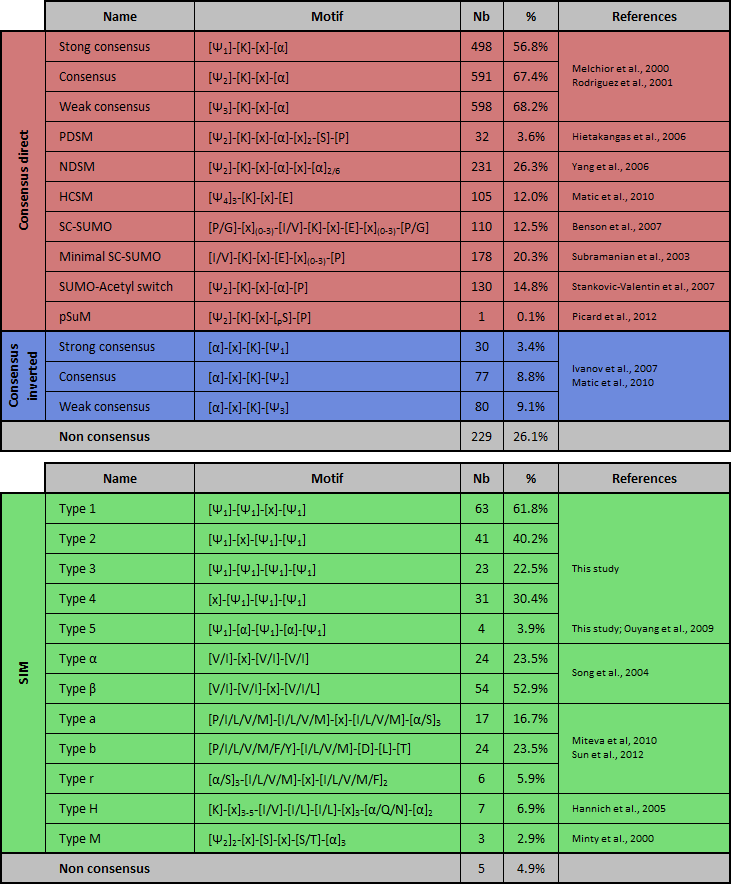 Table 1. List of known covalent or non-covalent SUMO-binding sites and abundance in the database of JASSA. PSDM: phosphorylation-dependent SUMOylation motif; NSDM: negatively charged amino acid-dependent SUMOylation motif; HCSM: hydrophobic cluster SUMOylation motif; SCM: synergy control motif; pSuM: phosphorylated SUMOylation motif. The abundance (number (Nb) and percentage (%)) of each motif in the DB is indicated. Ψ1 = I,L, or V; Ψ2 = A,F,I,L,M,P,V or W; Ψ3 = A,F,G,I,L,M,P,V,W or Y; Ψ4 = A,F,G,I,L,P or V; α = D or E; h = hydrobobic amino acid; pS/T = phosphorylated serine/threonine; SIM: SUMO interacting motif. ND: not determined. About JASSA
Comming soon... In update...
Lists of motifs used in JASSA
• List of SUMOylated lysines
• List of SIMs • List of SUMOylated and non SUMOylated lysines used for testing (Chen et al., 2012) • List of SIMs and non SIMs used for testing Citation
For publication of results, please cite the following article:
JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs Beauclair G, Bridier-Nahmias A, Zagury JF, Saïb A, Zamborlini A. Bioinformatics. 2015 Nov 1;31(21):3483-91. doi: 10.1093/bioinformatics/btv403. Epub 2015 Jul 2. PMID: 26142185 Update
• Update of text and picture (2015/02/10)
• Add "Best Prediction" setup (2015/01/10) • Add exportation of results in PyMol files format (.pml) (2014/07/29) • Update detection of Acidic/Serine stretch (2014/03/12) • Add predictive score (PS) to SIM analyse (2014/03/10) • Add Database Hit (DB Hit) to SIM analyse (2014/03/10) • Add detection of new SIM (Type 1 to 5) (2014/02/23) • New design of the output (2014/02/23) • Add and update of "Help" popup (2014/01/18) • Update of text and picture (2013/09/08) • Update of "About SUMOylation" (text and picture) (2013/09/06) • Add detection of SIM-a, SIM-b and SIM-r (2013/09/03) • Add detection of Acidic/Serine stretch (2013/07/25) • Add highlight of serine or acid amino acid in the environment (2013/07/25) • Add detection of pSuM and SUMO-Acetyl switch (2013/07/25) • ... • Creation of JASSA for SUMO site and SIM predictions (2010/01/19 03:12 AM ;p ) |